Despite the recent advancements in technology and chemical instrumentation, it is not yet feasible to measure cross sections for all sequences of small peptides. Consider, for example, five-residue tryptic fragments containing a C-terminal arginine or lysine residue; there will be 320,000 [2(20)4] different sequences. A method for accurate prediction of cross sections would be valuable. For the purposes of this discussion, a prediction is considered to be accurate if it falls within 2% of the experimental values, as this is often the level of accuracy reported for ion mobility measurements of cross section [Pub. 39].
We have developed an instrumental-based method for quickly determining average cross sections for components of complex mixtures of peptides or digests of large proteins. This has allowed for the creation of a database of nearly 3000 singly and doubly charged peptide ions obtained from the tryptic digestion of common proteins. Prior to the completion of this database, cross sections for only a few dozen peptides had been reported. As the cross section can provide valuable information about the three-dimensional structure of the ions, a large and actively updated collection of these measurements should benefit efforts to understand the intrinsic structural properties of these peptides in the absence of solvent and perhaps illuminate factors that influence folding.
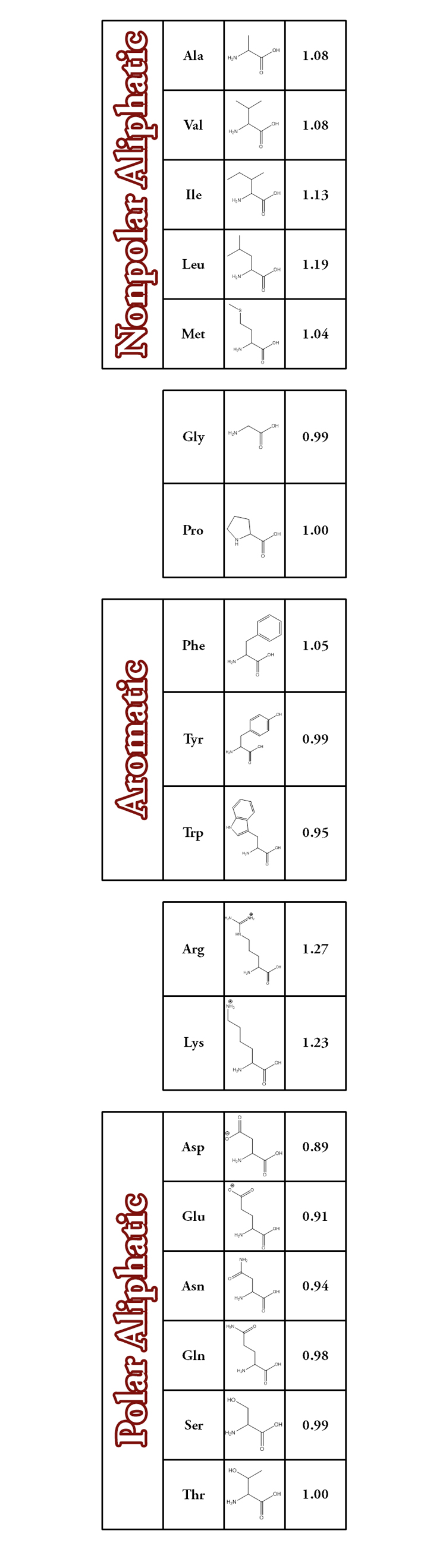
From the original database containing nearly 700 different peptide ions, a subset of more than 100 lysine-terminated peptide sequences that do not contain additional Lys, Arg, His, or Cys was created. Analysis of these related peptide sequences has shown that there are correlations of measure cross section with the amino acid composition. The influence of each amino acid on the observed cross section of the peptide has been quantified in terms of an intrinsic size parameter by solving for a system of equations that relates the frequency of the amino acid to measured cross section for different peptide sequences in the array of data. This was accomplished by writing a large system of many equations relating the occurrence frequency and unknown amino acid sizes to a reduced cross section of each peptide as given by the following equation.
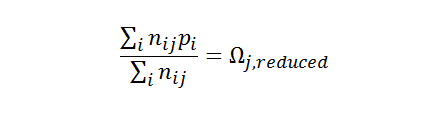
As shown here, nij represents the number of times an amino acid i occurs in a given sequence j, Ωj,reduced is the reduced cross section of sequence j, and pi is the unknown amino acid size parameter. The array of equations can be solved for the average, best-fit intrinsic size parameters (pi) using a linear algebra regression method. Using this method, more than 90% of the calculated cross sections were within 2% of the experimentally-obtained values.
Recently, we have expanded this original database of protonated species by adding alkali metals. For this study, nearly 1800 (~3x that of the original work) collision cross sections were obtained from the tryptic digest of 24 proteins. Here, such a large number of values enables generalizations on the assessment of peptide ion conformation resulting from solvation effects of the alkali cation or cations. For singly-charged and doubly-charged peptide ions, the cross section is generally observed to increase with increasing cationic size. Ultimately, we find that amino acid composition plays a significant role in the observed peptide ion cross sections.
These predictions allow us to better estimate data collected from the Waters SYNAPT instruments and thus increase our confidence in peak assignments from such experiments. Furthermore, this also allows us to look at other peptides with the same mass, but different composition. Lastly, as these datasets are almost impressively massive in terms of scale and the number of individual sequences represented, a large amount of work is being done in order to tease out structural variety between sequences and experimental conditions. For more information about these projects, please refer to the following publications, all of which are also available through the Publications page.