Clemmer Group
Ubiquitin Conformational Transitions
Protein Conformations Emerging from Solution into the Gas Phase
Ubiquitin [M+8H]8+ Ions (Native) Simulated at 470 K
Parameters
For the purposes of these ubiquitin coformational dynamics simulations, GROMACS (v 4.5.5) was used with the OPLS-AA force field. The native state structure was obtained from the Protein Data Bank (1UBQ). Additional conformations (A state and helix) were constructed using INSIGHT II.
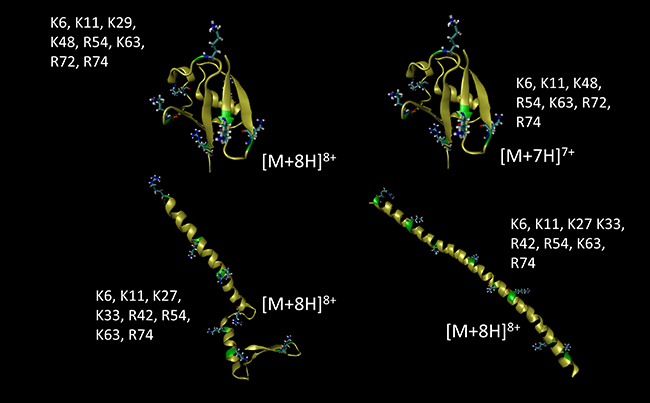
Charge Site Assignment
Ubiquitin has a large number of charge-carrying sites within the amino acid sequence. For the purposes of these calculations and simulations, the following charge-site configurations were employed.
Steps
- Energy Minimization (Steepest descent minimization)
- Simulated at 300 K for 50 ps
- Production Simulation:
- Temperature coupling (V-rescale)
- No pressure coupling
- No cutoff for electrostatics and van der Waals interactions
Selected References
- Berendsen, H. J. C., van der Spoel, D., and van Drunen, R. GROMACS: A Message-Passing Parallel Molecular Dynamics
Implementation. Comput. Phys. Commun. 1995, 91, 43–56.
- Lindahl, E., Hess, B. A., and van der Spoel, D. GROMACS 3.0: A Package for Molecular Simulations and Trajectory Analysis,
J. Mol. Model. 2001, 7, 306–317.
- van der Spoel, D., Lindahl, E., Hess, B., Groenhof, G., Mark, A. E., and Berendsen, H. J. C. GROMACS: Fast, Flexible and Free, J. Comput. Chem. 2005, 26, 1701–1718.
- Hess, B.; Kutzner, C.; van der Spoel, D.; Lindahl, E.; GROMACS 4: Algorithms for Highly Efficient, Loaded-Balanced, and Scalable Molecular Simulation. J. Chem. Theory Comput. 2008, 4, 435–447.
- Miteva, M.; Demirev, P. A.; Karshikoff, A. D. Multiply-Protonated Protein Ions in the Gas-Phase: Calculation of the Electrostatic Interactions between Charged Sites. J. Phys. Chem. B 1997, 101, 9645–9650.
- Breuker, K. Oh, H.; Horn, D. M.; Cerda, B. A.; McLafferty, F. W. Detailed Unfolding and Folding of Gaseous Ubiquitin Ions Characterized by Electron Capture Dissociation. J. Am. Chem. Soc. 2002, 124, 6407–6420.